My Google Summer of Code with Sktime - Neuroscience Meets Data Science
final project report
Introduction
For the past 10 weeks, I’ve been a Student Developer at sktime working 18 hours per week on tools to encourage the reproducibility of neuroscientific research. sktime is a unified interface for machine learning with time series that includes forecasting, data transformers, and classification and clustering algorithms. In this Google Summer of Code project, we created a sister package sktime-neuro with pre-processing strategies typically used prior to classifying EEG data. By creating an end-to-end pipeline in python that entails basic pre-processing algorithms and classification approaches we aim to encourage reproducible research in the field and facilitate more widespread analysis.
Some Background
EEG Measures Electrical Activity of the Brain!
Brains are fascinating. Billions of neurons are connected via synapses communicating with small electric currents, thereby orchestrating your movements, speech, thoughts, even your dreams, and much more. Yet our knowledge of the exact functioning of the brain is extremely limited. Electroencephalography (EEG) helps us to gain insights on this: The burst of a single neuron cannot be reliably detected by an EEG device, but whenever thousands of neurons fire synchronously they create an electrical field that spreads through our brain tissue, bone, and skull and can eventually be measured by placing electrodes on our head.
Finding the Signal Buried in Noise
As you can imagine the measured signal that has traveled through brain tissue, bone, and skull prior to being captured at the electrodes is pretty noisy. Subjects and investigators need to show endurance: the smallest movements, such as eye movements or clenching your teeth, will be visible as artifacts in your recordings. Even sweating can affect the conductance and may lead to electrode drift. (Many more biological and technical artifacts occur, for an overview see this blogpost [1])
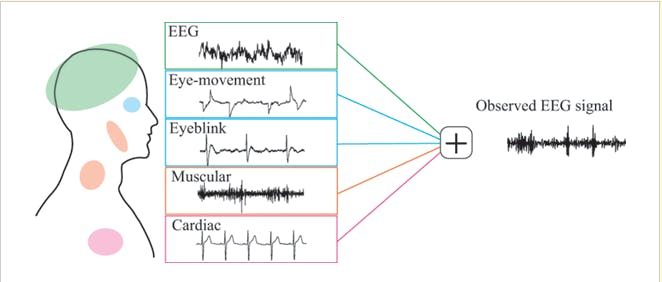 Figure 1. Examples of common EEG artifacts. Image is taken from [2].
Figure 1. Examples of common EEG artifacts. Image is taken from [2].
Pre-Processing Strategies are not Well Standardized
Different pre-processing strategies exist to mitigate the effects of those artifacts and to increase the signal-to-noise ratio: You may detrend your data to get rid of electrode drift, filter your data to remove slow frequency noise, or just detect and reject bad segments of data. However, these pre-processing strategies are not well standardized. Maaany different approaches exist ([3],[4],[5],[6] to name just a few) and even studies that follow a recommended pipeline still introduce their own variations of these pipelines [7]. Little is known on how sensitive EEG data analysis results are to those variations in pre-processing [7]. The goal of this project was to provide tooling to easily create and evaluate EEG processing pipelines for classification.
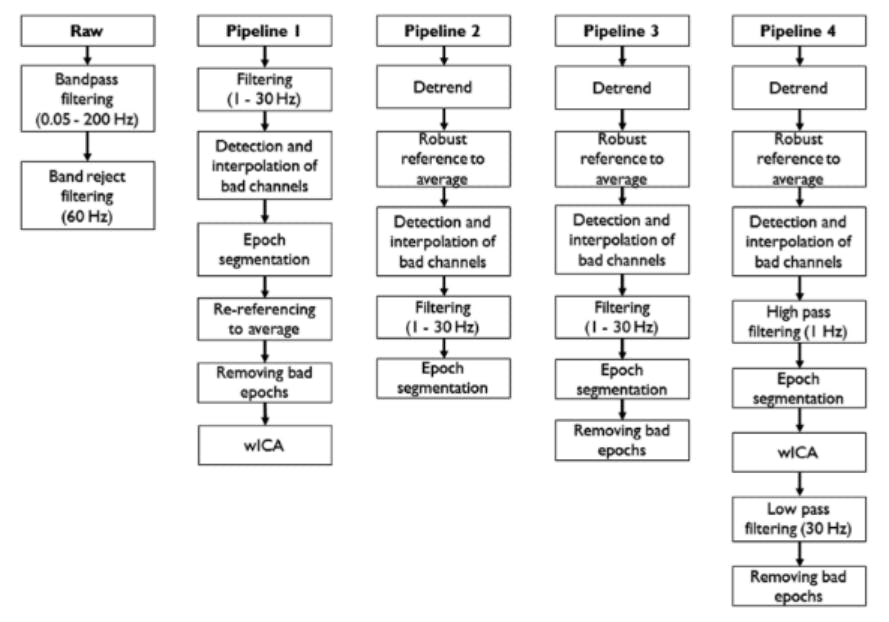 Figure 2. Examples of EEG processing pipelines. Taken from [8].
Figure 2. Examples of EEG processing pipelines. Taken from [8].
My Google Summer of Code Project
Sktime already provides many tools for classifying time series data (such as EEG data), including time series transformers that for example detrend the data. As part of this GSoC Project, we introduced a new sister package of sktime: sktime-neuro !
The concrete goals were:
- to provide functionality for some further basic EEG pre-processing strategies in the form of series-to-series and panel-to-panel transformers that can be used (or left out) interchangeably.
- to thereby provide an easy way to evaluate new processing algorithms/pipelines.
- to do some first basic benchmarking using the created pipeline on a set of Motor Imagery classification problems.
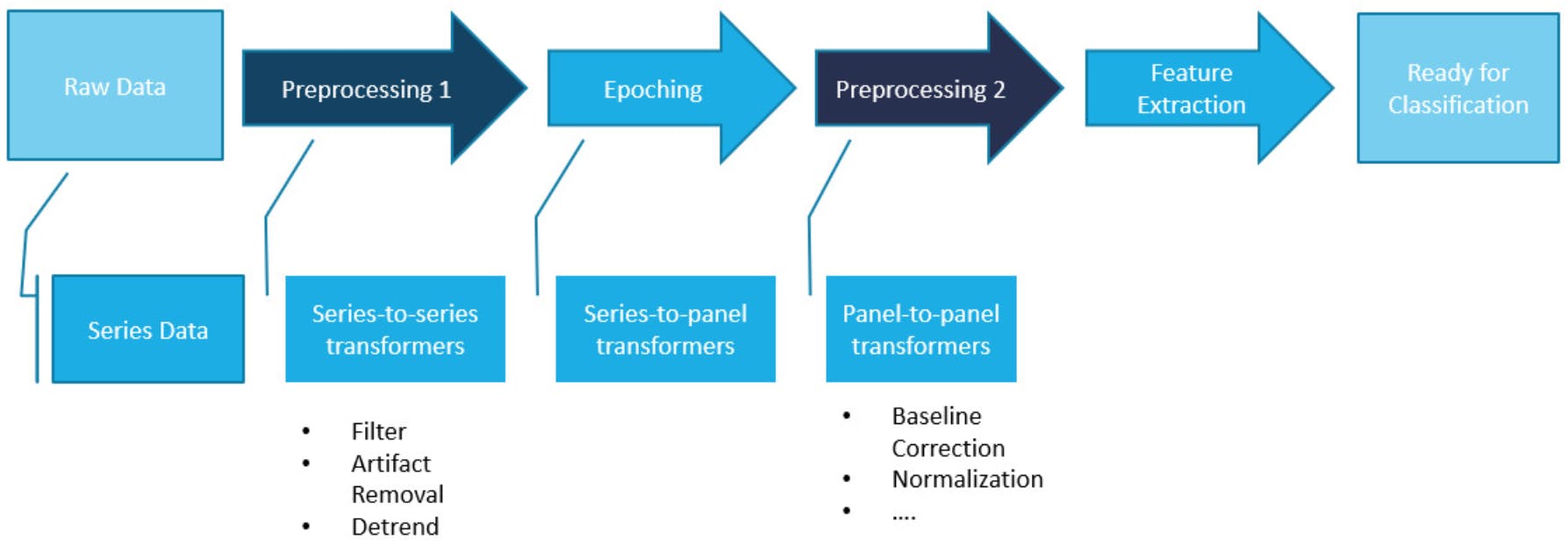
Figure 3. Visualization of the project idea. Pre-epoching pre-processing implemented as Series-to-series transformers. Post-epoching pre-processing implemented as Panel-to-panel transformers.
Code Contributions
1. Allowing sktime transformers for univariate series to be applied to multivariate series
2. Minor contributions on setting up the new repository
3. Panel-to-Panel and Series-to-Series Transformers as EEG Pre-Processing Strategies
4. Epoching
5. Example Notebook on matching pennies dataset
6. Artifact Removal (only experimental, challenging, only included in draft)
7. Contributions to Benchmarking
Becoming Part of the sktime Community
Part of my GSoC was also joining an incredibly welcoming and encouraging community. At the beginning of my internship, onboarding days for newcomers helped me to get started. After some weeks we gathered for a dev-sprint, where I gave a midterm presentation about my project and was involved in our roadmap planning. I also joined the community team with which we prepared a docsprint and applied to present sktime at PyData Global. I really enjoyed being part of this community and would like to thank the outreachy interns Guzal and Taiwo and our mentors Tony, Martina, Franz, and Markus as well as the entire sktime community for creating such a warm and supportive environment.
Sources
[1] bitbrain.com/blog/eeg-artifacts
[2] Kanoga, S., & Mitsukura, Y. (2017). Review of artifact rejection methods for electroencephalographic systems. Electroencephalography, 69(Nov), 69-89.
[3] Gabard-Durnam, L. J., Mendez Leal, A. S., Wilkinson, C. L., & Levin, A. R. (2018). The Harvard Automated Processing Pipeline for Electroencephalography (HAPPE): standardized processing software for developmental and high-artifact data. Frontiers in neuroscience, 12, 97.
[4] Bigdely-Shamlo, N., Mullen, T., Kothe, C., Su, K. M., & Robbins, K. A. (2015). The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Frontiers in neuroinformatics, 9, 16.
[5] Debnath, R., Buzzell, G. A., Morales, S., Bowers, M. E., Leach, S. C., & Fox, N. A. (2020). The Maryland analysis of developmental EEG (MADE) pipeline. Psychophysiology, 57(6), e13580.
[6] da Cruz, J. R., Chicherov, V., Herzog, M. H., & Figueiredo, P. (2018). An automatic pre-processing pipeline for EEG analysis (APP) based on robust statistics. Clinical Neurophysiology, 129(7), 1427-1437.
[7] Robbins, K. A., Touryan, J., Mullen, T., Kothe, C., & Bigdely-Shamlo, N. (2020). How sensitive are EEG results to preprocessing methods: a benchmarking study. IEEE transactions on neural systems and rehabilitation engineering, 28(5), 1081-1090.
[8] Suárez-Revelo, J. X., Ochoa-Gómez, J. F., & Tobón-Quintero, C. A. (2018, October). Validation of EEG pre-processing pipeline by test-retest reliability. In Workshop on Engineering Applications (pp. 290-299). Springer, Cham.

